Funtional Brain ROI Clustering with InfoMap

My first project at Banks Lab, in the Department of Anesthesiology at UW-Madison, involved utilizing InfoMap to achieve a functional clustering of regions of interest (ROIs) within the brain. InfoMap employs a random walk in a Markov model to cluster the model based on its network flow. Imagine taking a random walk along our Markov model. Afterward, we use Huffman coding to assign each node a binary name based on the frequency of visitation. We then use these new names to record the path in which we visited our nodes. InfoMap aims to compress this description of our journey without losing any information, thereby clustering our nodes in a functional manner. This is achieved by grouping nodes into so-called ‘cities.’ Each city has a unique name, and each node within a city has a unique name. However, nodes in different cities can share names, similar to how we can use Seattle and San Francisco as modifiers to differentiate between their respective Maple streets. We can take this idea further by considering how a state is a modifier for a city and a country is a modifier for a city, and so on. Now, with cities, we don’t just record when we visit a node, but also when we enter and leave a city. Clustering nodes into cities reduces our journey’s description length while clustering our nodes, accomplishing two goals simultaneously. For a deeper general overview, check out this Towards Data Science post or The Map Equation paper.
There are various hyperparameters that we can adjust to influence more or fewer clusters. The most pertinent of these is Markov Time, which allows you to control the link flow to change the cost of moving between cities (lower means more cities, higher means fewer). These hyperparameters were useful since our data was densely connected. Under default settings, all nodes would be clustered into a single city. Lowering the Markov time helps discourage a single cluster, revealing more interesting results, as seen below.
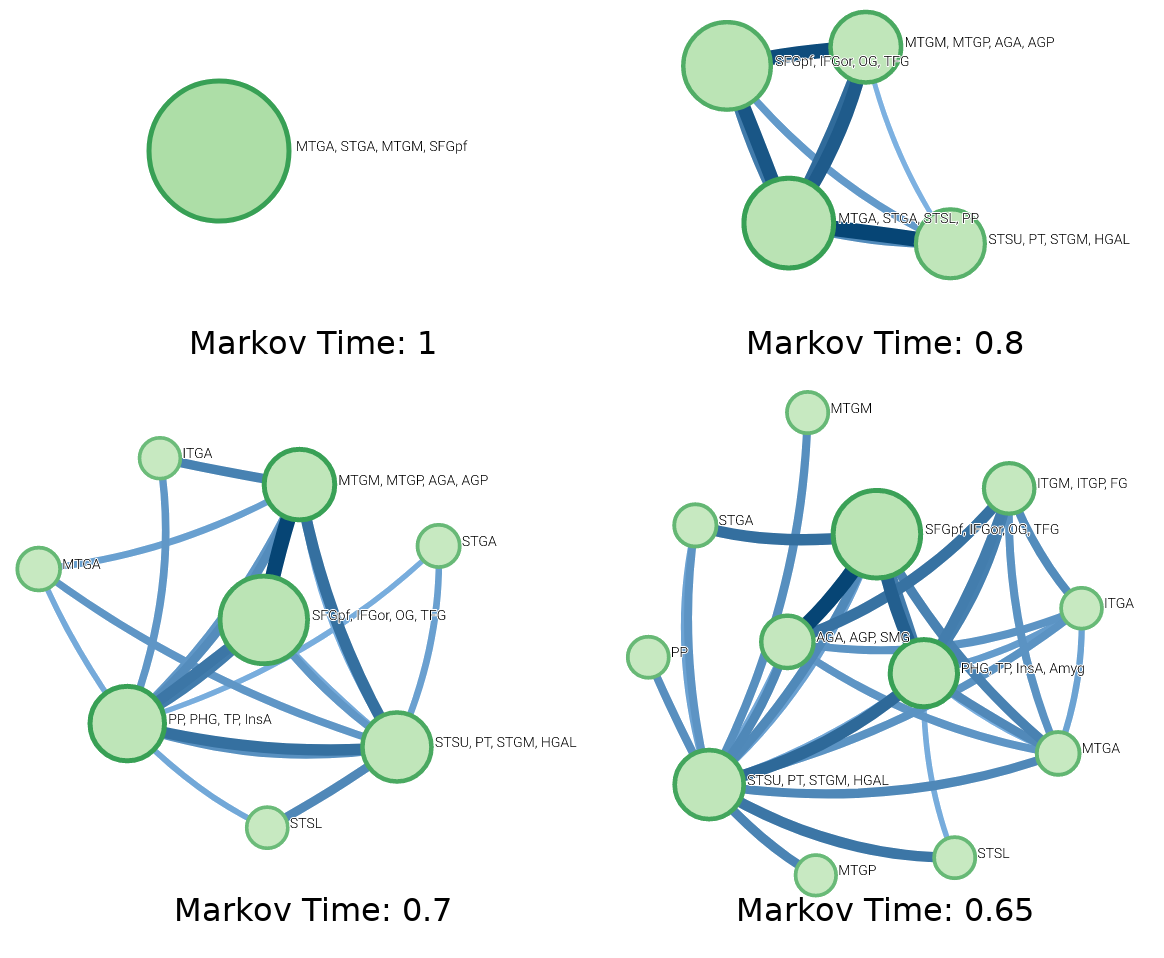
I applied InfoMap to intracranial electrophysiological data collected from epilepsy patients and used various connectivity measures on our electrode time series, which were then used as transition probabilities for my Markov models. These results are from the second attempt at using InfoMap. The first attempt yielded no significant results, but it taught me a valuable lesson: Before starting a project, read all relevant research, even if you don’t think it will be useful. Critically questioning your results and having a second pair of eyes are essential. After my first failure, I returned to InfoMap because my Principal Investigator, Matt Banks, obtained some interesting results clustering after applying Diffusion Map Embedding to his dataset. I wondered if I could achieve comparable results, and I did. I don’t know if these results will ever be published, but it’s nice to find closure on my first failed project.